The Matlab environment¶
Overview¶
Matlab can be used on the cluster in several configurations.
- run jobs directly on the compute nodes of the cluster (recommended)
out of the box parallelism up to 64 cores (a full max size node)
full parallelism on the cluster (guide not available yet)
- run jobs on a client and use the cluster as a backend (requires setup).
supports windows clients
supports linux and mac clients
Matlab as a client on Windows¶
This configuration allows the user to use MATLAB on a local machine e.g. a
laptop or a terminal on the AUB network and run the heavy computations sections
of a Matlab program/script on the HPC cluster. After the execution on the
HPC cluster is complete the results are transparently retrieved by MATLAB
and shown in the matlab workspace on the client. For this use case, the user
does not have to login (or interact) with the HPC cluster.
Note
this section of the guide has been tested with Matlab 2019b make sure you have the same version on the client machine.
Note
Multiple such parallel configuration can co-exist and can be selected at runtime.
Setting up a Matlab 2019b client¶
Pre-requisites:
Matlab 2019b installed on the client.
slurm.zip folder to be extracted in the integration folder
A working directory (folder) on your “C” or “D” drive.
Have your Matlab code modified to exploit parallelism.
Once
slurm.zipis downloaded, extract it toDocuments\MATLAB(shown in screenshot below) or to the corresponding directory of your non-default Matlab installation directory:Open Matlab R2019b on the client machine (e.g your laptop)
Select
Set Path(under HOME -> ENVIRONMENT)Click on
Add FolderBrowse to
Documents\MATLAB\slurm\nonsharedClick save
To import the
octopus.mlsettingsprofile:click on
Parallelclick on
Manage Cluster Profiles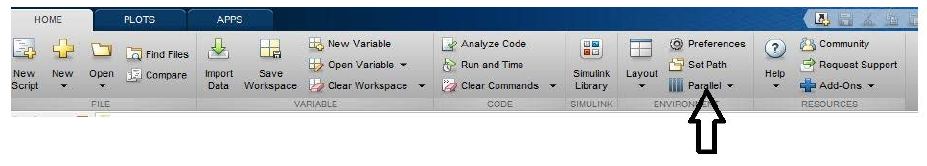
Choose
Importthen browse tooctopus.mlsettingsfile (downloaded in step 3 in the Pre-requisites section above)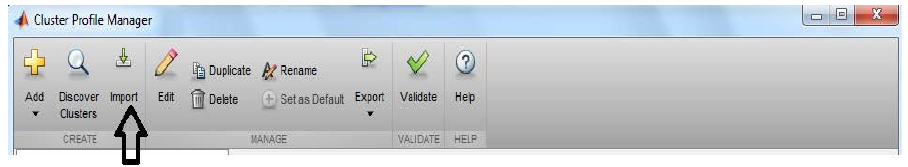
Once the
octopus.mlsettingsprofile gets loaded, select it, click onEdit, and modify theRemoteJobStorageLocationby using a path on your HPC account (make sure to change the<user>to your username).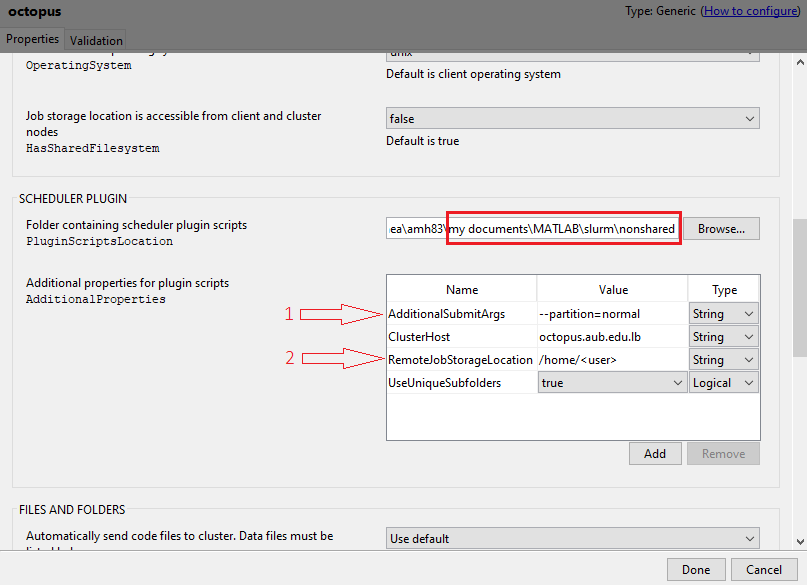
You can choose which queue to work on through modifying
AdditionalSubmitArgs:You can modify the number of cores to be used on HPC cluster (e.g. 4,6,8,10,12) through
NumWorkers
When finished, press done and make sure to set the HPC profile as
Default.Press
validateto validate the parallel configuratin.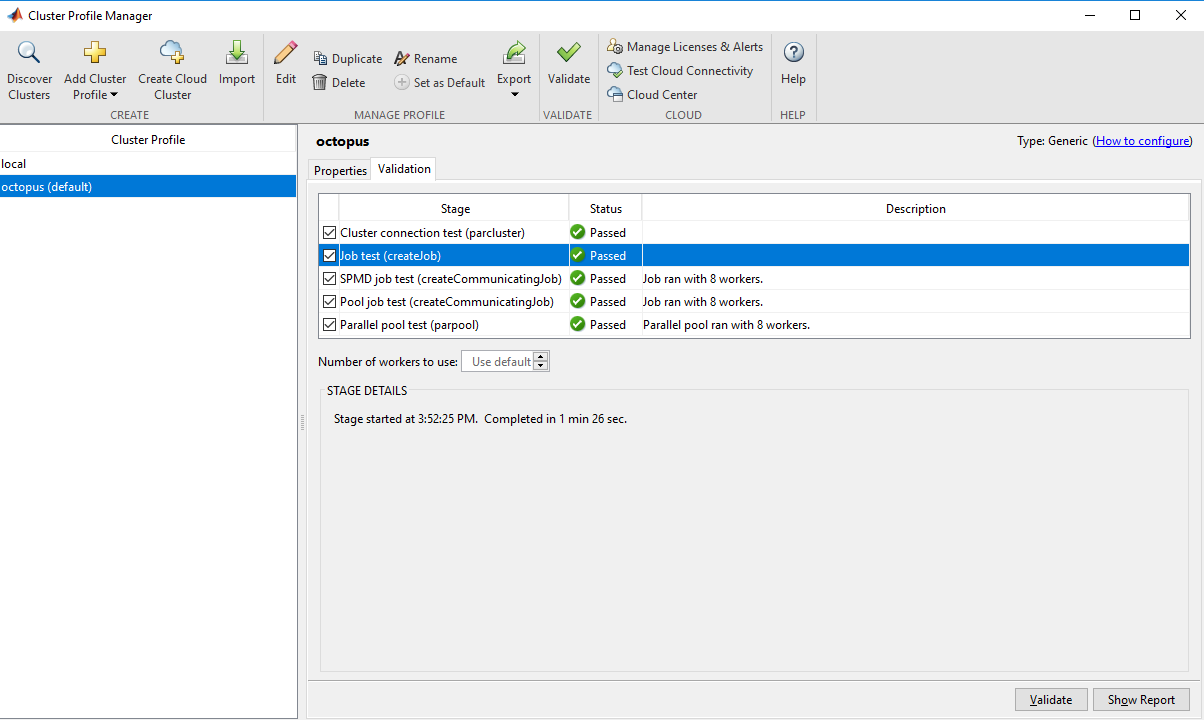
Client batch job example¶
Below is a sample Matlab program for
submitting independent jobs on the cluster. In this script four functions are
exectued on the cluster and the results are collected back one job a time back
to back in blocking mode (this can be improved on but that is beyond the scope
of this guide).
clc; clear;
% run a function locally
output_local = my_linalg_function(80, 300);
% run 4 jobs on the cluster, wait for the remote jobs to finish
% and fetch the results.
cluster = parcluster('Octopus');
% run the jobs (asyncroneously)
for i=1:4
jobs(i) = batch(cluster, @my_linalg_function, 1, {80, 600});
end
% wait for the jobs to finish
for i=1:4
status = wait(jobs(i));
outputs(i) = fetchOutputs(jobs(i));
end
% define a function that does some linaer algebra
function results = my_linalg_function(n_iters, mat_sz)
results = zeros(n_iters, 1);
for i = 1:n_iters
results(i) = max(abs(eig(rand(mat_sz))));
end
end
Note
Fetching outputs will fail if more than one instance of Matlab is connecting to the cluster for that user. So two Matlab instances on the same client or two Matlab instances on two different clients (one on each client) will cause the synchronization of job results with SLURM to fail. to correct this, you must change the JobStorageLocation in the cluster profile (the local folder to which jobs are synched)
Note
For communicating jobs using shared memory or MPI the jobs should be submitted on the cluster directly and it is not possible to submit such jobs through the client in the configuration described above.
Matlab on the compute nodes of the cluster¶
This configuration allows the user to run MATLAB scripts on the HPC cluster directly through the scheduler. Once the jobs are complete the user can choose to transfer the results to a local machine and analyze them or analyze everything on the cluster as well and e.g retrieve a final product that could be a plot or some data files. This setup does not require the user to have matlab installed on their local machine.
Serial jobs¶
No setup is required to run a serial job on the cluster.
The following job script (matlab_serial.sh) can be used to submit a serial job
running the matlab script my_serial_script.m.
#!/bin/bash
#SBATCH --job-name=matlab-smp
#SBATCH --partition=normal
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=1
#SBATCH --cpus-per-task=1
#SBATCH --mem=16000
#SBATCH --time=0-01:00:00
module load matlab/2018b
matlab -nodisplay -r "run('my_smp_script.m'); exit" > matlab_${SLURM_JOBID}.out
tic
values = zeros(200);
for i = 1:size(values, 2)
values(i) = sum(abs(eig(rand(800))));
end
toc
disp(sum(sum(values)));
The following should be present in the output
Elapsed time is 113.542701 seconds.
checksum = 9.492791e+05
Note
the Elapsed time could vary slightly since the execution time
depends on the load of the compute node (if it is not the only running process)
and the checksum could vary slightly since it is based on randon numbers.